Welcome to 14-3-3.com, a website dedicated to disseminating information about 14-3-3 proteins.
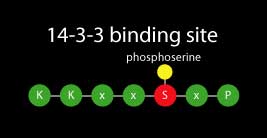
14-3-3s are soluble dimeric proteins that were originally named for their migration behavior in a laboratory assay, however today they are recognized as major regulatory proteins involved in many physiological processes of all studied eukaryotic organisms. 14-3-3s alter function of other proteins by binding predominantly to a phosphorylated serine or threonine residue located in a conserved motif within the target protein.
The identification of 14-3-3 target proteins was originally determined one protein at a time. Today however, large scale proteomic 14-3-3 capture experiments coupled with mass spectrometry can reveal 100's of protein targets with a single experiment. While a vast improvement over single target detection and start to address the widescale biological impact of 14-3-3s, these processes still only identify a sampling of all 14-3-3 targets.
With the completion of entire genome seqeuncing projects and using bioinformatics it is possible to predict genome-wide potential 14-3-3 targets for a given organism based upon the conserved binding motif.
Analysis of potential and experimentally confirmed targets for entire and or multiple genomes should help further the 14-3-3 research field. An example of such analysis is provided below for the plant molecular biology model system Arabidopsis.
Searchable database of 14-3-3 binding Arabidopsis proteins

Searchable database of all Arabidopsis proteins
